The West Nile virus invasion
West Nile virus was first detected in the United States in 1999 in New York, and within a few years, the virus spread across the country. Now, West Nile virus is endemic and has become the most important mosquito-borne virus threat in the United States. Several countries in Europe and the Americas have also recently started observing West Nile outbreaks. Yet, little progress has been made in controlling outbreaks, due in part to a declining interest in West Nile virus research. Thus, for effective targeted control measures to be developed, there is a critical need to explore the diversity of circulating West Nile viruses and how this may influence the emergence of new virus strains that cause disease outbreaks.
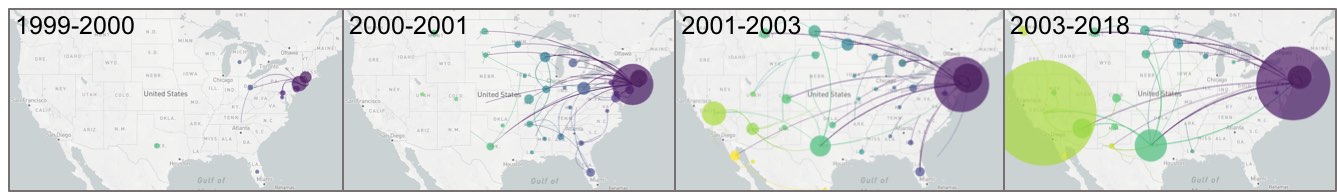
High-definition West Nile virus phylogenetics
The WestNile 4K Project is creating a large resource by partnering public health laboratories and academic institutions to perform phylogenetic and experimental studies aimed at answering fundamental questions about West Nile virus ecology, epidemiology, and evolution across the United States.
- How far does West Nile virus currently spread? What are the barriers?
- Are West Nile virus outbreaks caused by newly introduced viruses or by long-lived lineages?
- Has West Nile virus adapted to regions within the United States?
- Do different circulating West Nile virus strains have different outbreak potential?
To answer these questions, plus many more, we need to fill in our many gaps that remain regarding West Nile virus diversity in the United States. Thus, our goal is to sequence at least 8,000 West Nile virus from across the country, focusing primarily on isolates since West Nile virus has become endemic in this country. To understand phenotypic consequences of viral diversity and investigate West Nile virus evolution, we will couple our genomic studies to extensive high-throughput experimental studies in vitro and in vivo.
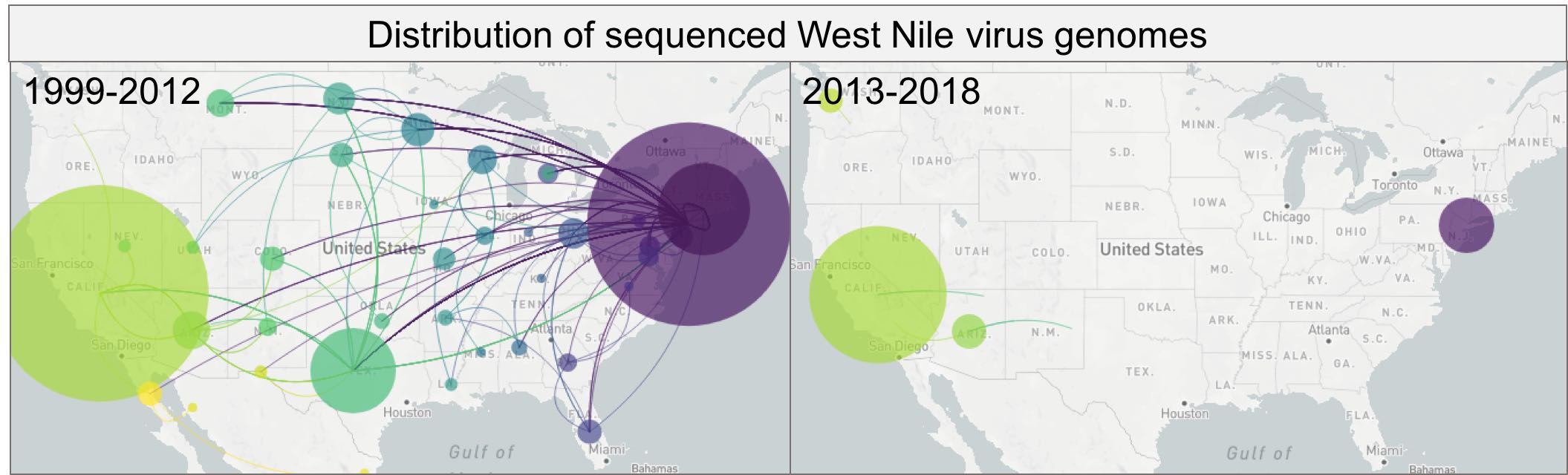
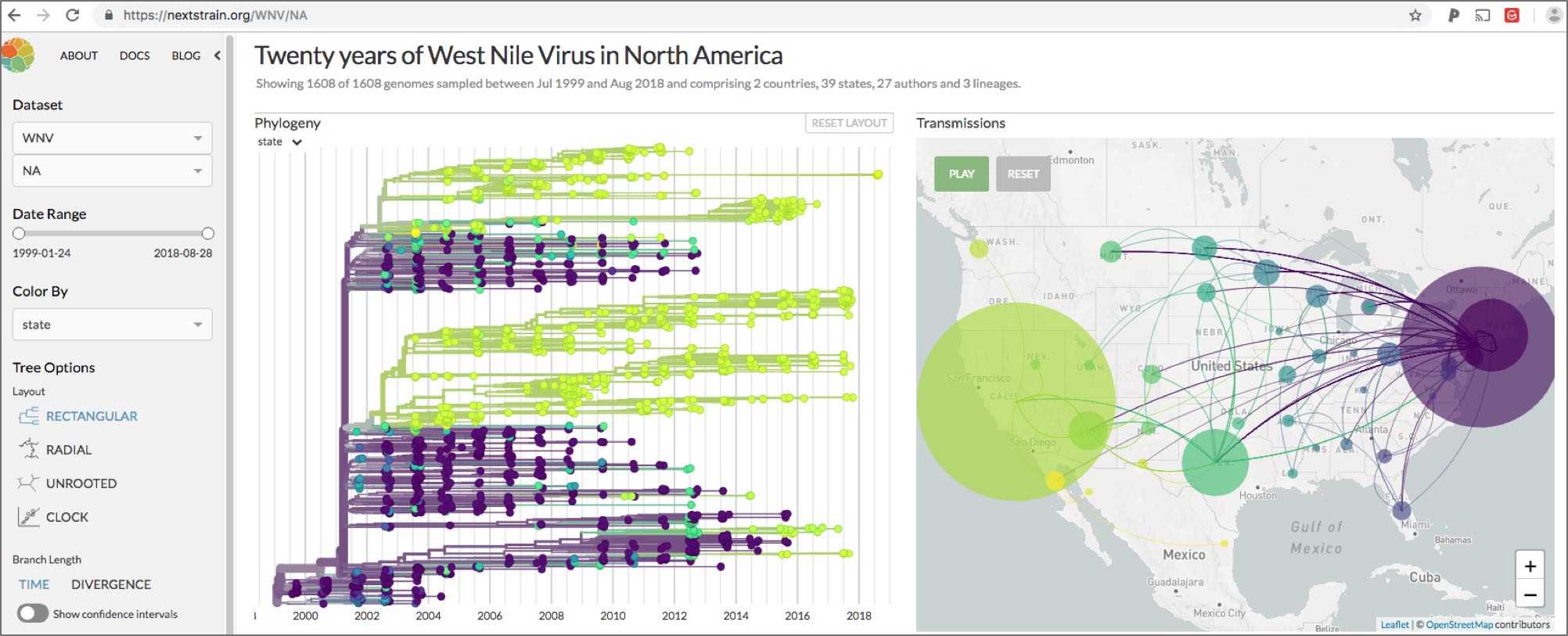
Open data sharing and visualization
The genomic data produced as part of the WestNile 4K Project is openly available immediately after generation (https://andersen-lab.com/secrets/data/west-nile-genomics-westnile-4k/) and our results are visualized using Nextstrain (https://nextstrain.org/WNV), an interactive online tool to visualize West Nile virus spread using genomic epidemiology.
We need your help
There are already dozens of collaborators joining us to better understand West Nile virus in the United States, but we need more help. Please let us know if you have a freezer full of West Nile virus samples from the last few years and/or if you plan on doing collections this year. By receiving samples from you, we will sequence virus genomes at no cost to you and share the resulting data directly with you and publicly via this website. You are free to use the generated data for your own studies and large collaborative studies and publications are planned with all collaborators directly included in study design and manuscript preparations.
If you are interested in joining the WestNile 4K Project and help increase our understanding of the spread and evolution of the virus, please reach out to use directly via the emails below. It would be helpful if you could please also provide the following information:
- Location of sample collection (state and county)
- Years in which you have West Nile virus samples
- The number of samples that you could provide
On behalf of the entire WestNile 4K Project, we would be thrilled to include you in this study, so please don’t hesitate to reach out to us if answer any questions or are interested in joining.
Nate – [email protected]
Ryan – [email protected]
Marc – [email protected]
Kristian – [email protected]
